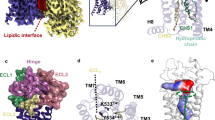
Abstract
Frizzled receptors (FZDs) are class-F G-protein-coupled receptors (GPCRs) that function in Wnt signalling and are essential for developing and adult organisms1,2. As central mediators in this complex signalling pathway, FZDs serve as gatekeeping proteins both for drug intervention and for the development of probes in basic and in therapeutic research. Here we present an atomic-resolution structure of the human Frizzled 4 receptor (FZD4) transmembrane domain in the absence of a bound ligand. The structure reveals an unusual transmembrane architecture in which helix VI is short and tightly packed, and is distinct from all other GPCR structures reported so far. Within this unique transmembrane fold is an extremely narrow and highly hydrophilic pocket that is not amenable to the binding of traditional GPCR ligands. We show that such a pocket is conserved across all FZDs, which may explain the long-standing difficulties in the development of ligands for these receptors. Molecular dynamics simulations on the microsecond timescale and mutational analysis uncovered two coupled, dynamic kinks located at helix VII that are involved in FZD4 activation. The stability of the structure in its ligand-free form, an unfavourable pocket for ligand binding and the two unusual kinks on helix VII suggest that FZDs may have evolved a novel ligand-recognition and activation mechanism that is distinct from that of other GPCRs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nusse, R. & Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Clevers, H. & Nusse, R. Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 (2012).
Dijksterhuis, J. P., Petersen, J. & Schulte, G. WNT/Frizzled signalling: receptor-ligand selectivity with focus on FZD-G protein signalling and its physiological relevance: IUPHAR Review 3. Br. J. Pharmacol. 171, 1195–1209 (2014).
Schulte, G. International union of basic and clinical pharmacology. LXXX. The class Frizzled receptors. Pharmacol. Rev. 62, 632–667 (2010).
Wang, Y., Chang, H., Rattner, A. & Nathans, J. Frizzled receptors in development and disease. Curr. Top. Dev. Biol. 117, 113–139 (2016).
Kahn, M. Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 13, 513–532 (2014).
Tao, L. et al. Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature 538, 350–355 (2016).
Nile, A. H., Mukund, S., Stanger, K., Wang, W. & Hannoush, R. N. Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding. Proc. Natl Acad. Sci. USA 114, 4147–4152 (2017).
DeBruine, Z. J. et al. Wnt5a promotes Frizzled-4 signalosome assembly by stabilizing cysteine-rich domain dimerization. Genes Dev. 31, 916–926 (2017).
Janda, C. Y., Waghray, D., Levin, A. M., Thomas, C. & Garcia, K. C. Structural basis of Wnt recognition by Frizzled. Science 337, 59–64 (2012).
Chang, T. H. et al. Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan. eLife 4, (2015).
Shen, G. et al. Structural basis of the Norrin–Frizzled 4 interaction. Cell Res. 25, 1078–1081 (2015).
Janda, C. Y. et al. Surrogate Wnt agonists that phenocopy canonical Wnt and β-catenin signalling. Nature 545, 234–237 (2017).
Gurney, A. et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc. Natl Acad. Sci. USA 109, 11717–11722 (2012).
Umbhauer, M. et al. The C-terminal cytoplasmic Lys-Thr-X-X-X-Trp motif in Frizzled receptors mediates Wnt/β-catenin signalling. EMBO J. 19, 4944–4954 (2000).
Byrne, E. F. X. et al. Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517–522 (2016).
Wang, C. et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature 497, 338–343 (2013).
Zhang, X. et al. Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand. Nat. Commun. 8, 15383 (2017).
Huang, P. et al. Structural basis of Smoothened activation in Hedgehog signaling. Cell 174, 312–324.e16 (2018).
Wang, Y. et al. Norrin/Frizzled4 signaling in retinal vascular development and blood brain barrier plasticity. Cell 151, 1332–1344 (2012).
Zhang, C. et al. Norrin-induced Frizzled4 endocytosis and endo-lysosomal trafficking control retinal angiogenesis and barrier function. Nat. Commun. 8, 16050 (2017).
Nikopoulos, K. et al. Overview of the mutation spectrum in familial exudative vitreoretinopathy and Norrie disease with identification of 21 novel variants in FZD4, LRP5, and NDP. Hum. Mutat. 31, 656–666 (2010).
Strakova, K. et al. The tyrosine Y2502.39 in Frizzled 4 defines a conserved motif important for structural integrity of the receptor and recruitment of Disheveled. Cell. Signal. 38, 85–96 (2017).
Zhang, H. et al. Structure of the full-length glucagon class B G-protein-coupled receptor. Nature 546, 259–264 (2017).
Jazayeri, A. et al. Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature 546, 254–258 (2017).
Wu, H. et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64 (2014).
Zheng, Y. et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature 540, 458–461 (2016).
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011).
Popov, P. et al. Computational design of thermostabilizing point mutations for G protein-coupled receptors. eLife 7, e34729 (2018).
Ma, Y. et al. Structural basis for apelin control of the human apelin receptor. Structure 25, 858–866.e4 (2017).
Cherezov, V. et al. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-µm size X-ray synchrotron beam. J. R. Soc. Interface 6, S587–S597 (2009).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D 68, 368–380 (2012).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Pau, M. S., Gao, S., Malbon, C. C., Wang, H. Y. & Bertalovitz, A. C. The intracellular loop 2 F328S Frizzled-4 mutation implicated in familial exudative vitreoretinopathy impairs Dishevelled recruitment. J. Mol. Signal. 10, 5 (2015).
Kramer, G. D., Say, E. A. & Shields, C. L. Simultaneous novel mutations of LRP5 and TSPAN12 in a case of familial exudative vitreoretinopathy. J. Pediatr. Ophthalmol. Strabismus 53, e1–e5 (2016).
Yuan, Y., Pei, J. & Lai, L. LigBuilder 2: a practical de novo drug design approach. J. Chem. Inf. Model. 51, 1083–1091 (2011).
Lomize, M. A., Lomize, A. L., Pogozheva, I. D. & Mosberg, H. I. OPM: Orientations of Proteins in Membranes database. Bioinformatics 22, 623–625 (2006).
Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).
MacKerell, A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998).
Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Eswar, N. et al. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 15, 5.6.1–5.6.30 (2006).
McGibbon, R. T. et al. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 109, 1528–1532 (2015).
Humphrey, W., Dalke, A. & Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graphics 14, 33–38 (1996).
Xu, T. H. et al. Alzheimer’s disease-associated mutations increase amyloid precursor protein resistance to γ-secretase cleavage and the Aβ42/Aβ40 ratio. Cell Discov. 2, 16026 (2016).
Yan, Y. et al. Dimerization of the transmembrane domain of amyloid precursor protein is determined by residues around the γ-secretase cleavage sites. J. Biol. Chem. 292, 15826–15837 (2017).
Chen, X. et al. A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline. Sci. Rep. 5, 8361 (2015).
Acknowledgements
This work was supported by the National Natural Science Foundation (NSF) of China grant 31670736 (F.X.), the National Key Research and Development Program of China grant 2018YFA0507004 (F.X.) and 2016YCF0905902 (S.Z.), the NSF of Shanghai grant 16ZR1448500 (S.Z.), the Russian Foundation for Basic Research (RFBR 18-34-00990) (P.P.) and Shanghai Municipal Government, ShanghaiTech University. The diffraction data were collected at BL41XU@Spring-8 with JASRI proposals 2016B2702. We thank J. Liu, X. Gu, N. Chen and L. Xue of the BV facility at the iHuman Institute, ShanghaiTech University for protein expression support; M. Hanson from the GPCR Consortium and K. Diederichs from the University of Konstanz for X-ray data processing; the mass spectrometry facility at the National Protein Science Center (Shanghai, China) for technical assistance; and A. Pautsch from Boehringer Ingelheim and W. Zhong from Amgen for discussions.
Reviewer information
Nature thanks M. Filizola, X. He, A. K. Shukla and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
S.Y. performed cloning, protein purification, crystallization, data collection, structural analysis and figure preparation; Y.W. carried out structure analysis, molecular dynamics simulations and figure preparation; T.-H.X. performed mutagenesis, cellular localization, cell surface biotinylation, and TOPflash reporter assays and corresponding figure preparation; P.W.d.W. carried out molecular dynamics simulations of ΔCRD-FZD4, structure analysis and corresponding figure preparation; Y.H., Z.J.D., Y.L., K.S.-P. and K.G.H. performed mutagenesis and TOPflash reporter assays; M.P. carried out molecular replacement and structure refinement; B.Z. carried out affinity mass spectrometry; S.A.Z. performed full-length modelling; P.P. was responsible for stabilizing mutation design; G.W.H. was responsible for structure refinement, quality control and deposition; Y.C. and S.D. performed cloning and protein purification; Y.G. carried out computational analysis; V.K. supervised the stabilization of mutation design and full-length modelling; W.S. supervised the affinity mass spectrometry analysis and table preparation; L.J.M., K.M. and H.E.X. designed cell-based experiments, data analysis and interpretations; R.C.S. supervised the structure analysis; S.Z. supervised the structure analysis, simulation and figure preparation; F.X. designed and supervised experiments and performed data analysis; and S.Y. and F.X. wrote the manuscript with discussions and improvements from Y.W., P.W.d.W., H.E.X., K.M., S.Z. and R.C.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Sequence alignment between FZD4 and nine other FZDs as well as SMO.
Colours represent the similarity of residues: red background, identical; red text, strongly similar. The alignment was generated using MAFFT (https://www.ebi.ac.uk/Tools/msa/mafft/) and the graphic was prepared on the ESPript 3.0 server (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
Extended Data Fig. 2 Crystallization of ΔCRD-FZD4 and structure determination.
a, Schematic of the ΔCRD-FZD4 construct. To obtain crystals that would diffract well, we truncated the N-terminal CRD region (residues 1–177) and C-terminal flexible region (residues 517–537) and introduced four single mutations (M309L, C450I, C507F and S508Y, coloured in red) that are designed based on sequence conservation analysis across ten human FZDs. Residues that are involved in the X.50f numbering system are coloured in green. The cysteines that form endogenous disulfide bonds are indicated in orange. b, Fluorescence-activated cell sorting staining data, to monitor the surface expression of the construct used in this study. The experiment was repeated twice with similar results. c, Crystals of ΔCRD-FZD4 in the apo state. d, Crystal packing of ΔCRD-FZD4.
Extended Data Fig. 3 Dynamics of helix V and VI in FZD4 and a comparison of the intracellular side of FZD4 with other GPCRs.
a, Cα r.m.s.d. for residues G409 to S418 (helix V) and R432 to S441 (helix VI) plotted as a rolling average over a 10-ns window. b, Unique hydrogen bonds between helix III and helix VI (W3203.43f–S4116.37f and W3273.50f–M4346.30f) maintain helix VI in an inward conformation. c, The intracellular cavity of FZD4 is close to W7.55f and the KTXXXW motif (coloured in dark grey), a region that is key for downstream signalling. Compared with class-A GPCRs (adenosine A2A and β2 adrenergic receptors), FZD4 leaves no cavity among helices III,VI and VII, whereas SMO has a side-pocket at this position. R3.50 (labelled with a blue sphere) is a key residue in the activation of class-A GPCRs and is exposed to the intracellular surface. The residue in the same position in class-F, W3.50f, points in a different direction and is not exposed. The G-protein-binding sites of class-A GPCRs are labelled in red.
Extended Data Fig. 4 Activity analysis of disease-related and signalling-related mutations of ΔCRD-FZD4, and the conformational change of family-conserved amino acids of ΔCRD-FZD4.
a, TOPflash analysis, cellular localization and cell surface expression analysis of disease-related and signalling-related mutations. Each data point represents mean ± s.e.m., repeated in triplicate. The experiment was repeated using two independent methods with similar results. All the mutations affected FZD4 downstream signalling in some way—some decreased the WNT3A–Norrin signal substantially, some showed gain-of-function activity—which suggests complex mechanisms underlying these mutation-caused diseases. b, Conformational rearrangement on family-conserved residues W4947.55f and Y2502.39f was observed when comparing FZD4 (cyan) with SMO (gold). It is noteworthy that Y2502.39f in FZD4 points outwards from the 7TM bundle.
Extended Data Fig. 5 The model of full-length FZD4 and molecular dynamics analysis.
a, The model of full-length FZD4. Cyan, TMD (crystal structure); magenta, linker; blue, CRD (modelled from PDB ID: 5CM4). The human smoothened receptor crystal structure (grey; PDB ID: 5L7D) is overlaid onto the model. Disulfide bridges in the FZD4 model are shown in stick representation. b, Motions of the CRD domain observed during the molecular dynamics simulation. Snapshots of the positions of the CRD domain and linker during one of three independent molecular dynamics simulations are shown in cartoon representation, at 250 ns (light blue), 500 ns (green), 750 ns (pink), 1,000 ns (yellow), 1,250 ns (orange) and 1,500 ns (red). The initial full-length CRD and linker domain are shown in blue, and all TMDs are shown in cyan. c, Motions of the CRD domain observed during the molecular dynamics simulation. The r.m.s.d. plot of Cα atoms of the CRD and linker domain with respect to the initial full-length FZD4 model during the three molecular dynamics simulations is shown.
Extended Data Fig. 6 Analysis of the TMD pocket of FZD4.
a, Superimposition of other SMO ligands (ANTA-XV, GDC0449, SAG, Cyclopamine and SANT-1) in the pocket of FZD4. All of these SMO ligands collide with the FZD4 pocket. b, The volume of the TMD pocket during simulation. Volumes are displayed as rolling averages over a 10-ns window. c, Total expression and cellular-localization analysis for pocket mutations. Total FZD–YFP expression is determined by YFP fluorescence; cellular localization is determined by fluorescence microscopy (related to Fig. 3). A plasmid expressing C-terminally sfGFP-tagged arrestin protein (Arr-ctrl), which localizes largely to the cytoplasm, was used as negative control for cell membrane localization. The experiment was repeated using two independent methods with similar results. d, The FZD4 pocket has the highest hydrophilic/hydrophobic ratio of all GPCR structures that have been solved to date.
Extended Data Fig. 7 Homology model and conservation analysis for ten human FZDs.
a, Superposition of ten human FZD homology models. The green, thin regions represents high conservation, and the red, thick regions represent low conservation. b, Sequence alignment of pocket residues, with conserved amino acids across the FZD family highlighted in dark green. c, Conserved amino acids in the top of the FZD4 pocket. d, Conserved amino acids in the bottom of the FZD4 pocket.
Extended Data Fig. 8 Molecular dynamics and mutation analysis of the Dishevelled-binding site.
a, The Dishevelled-binding site plotted as a moving average over a 10-ns window. Water molecules less than 10 Å from H348(N) were defined as proximal for this analysis. b, Total expression and cellular localization analysis for mutations in the Dishevelled-binding site (related to Fig. 4). The experiment was repeated using two independent methods with similar results.
Extended Data Fig. 9 Analysis of the two unusual kinks.
a, Two kinks with conserved polar networks fluctuate between bent and straight conformations during simulation. b, Molecular dynamics traces of the kink 1 and kink 2 backbone distances plotted as a moving average over a 10-ns window. c, Total expression and cellular localization analysis of kink 1/kink 2 mutations (related to Fig. 4). The experiment was repeated using two independent methods with similar results.
Supplementary information
Rights and permissions
About this article
Cite this article
Yang, S., Wu, Y., Xu, TH. et al. Crystal structure of the Frizzled 4 receptor in a ligand-free state. Nature 560, 666–670 (2018). https://doi.org/10.1038/s41586-018-0447-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0447-x